My research focuses on bridging coarse-grained and atomistic simulations using machine-learning–based probabilistic backmapping. I develop generative models that produce thermodynamically meaningful atomistic ensembles from coarse-grained configurations, enabling direct connection between efficient CG sampling and atomistic physics.
Generative models for one-to-many mappings
Ensemble recovery via reweighting / stabilization
Proteins & assemblies (IDPs, aggregation)
Soft matter, nucleation, porous materials
Current Focus
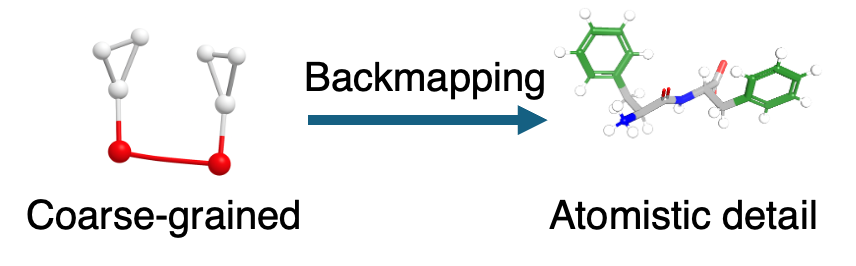
Probabilistic CG→AA backmapping with generative models
Using diphenylalanine as a model system, I develop
probabilistic normalizing-flow–based decoders that generate
ensembles of atomistic structures consistent with a given coarse-grained configuration.
The models capture conformational diversity, preserve correlations between internal degrees
of freedom, and produce physically realistic structures that can be used directly in
molecular dynamics simulations.
Ongoing work focuses on reweighting generated ensembles to recover
Boltzmann statistics, enabling quantitative study of
peptide self-assembly at atomistic resolution while retaining the efficiency of
coarse-grained sampling.
Stabilizing importance weights (higher effective sample size)
Because backmapping is intrinsically a high-variance importance-sampling problem,
practical reweighting can suffer from weight collapse.
I investigate stabilization strategies that improve effective sample size (ESS)
while preserving unbiased thermodynamic observables.
Research Themes
Proteins & intrinsically disordered systems
Sequence–ensemble relationships, aggregation propensity, and multiscale structure generation for IDPs and assemblies.
Chromatin / protein–DNA organization
Condensate and chromatin compaction dynamics from simulation, with emphasis on mechanistic, interpretable physical drivers.
Self-assembly, nucleation, porous materials
Mesophase self-assembly, polymorph selection, and nucleation pathways in nanoparticle and porous-material systems.
Methods Toolkit
Machine Learning
- Normalizing flows / diffusion-style score models (generative decoders)
- Conditional generation with calibrated probabilities
- Evaluation via reweighting, ESS, and observable agreement
Simulation & Analysis
- Molecular dynamics, coarse-graining, internal-coordinate representations
- Statistical mechanics, free energies, reweighting diagnostics
- Reproducible pipelines for training + generation + analysis
Selected Outputs
See Publications and Talks & Posters for details.